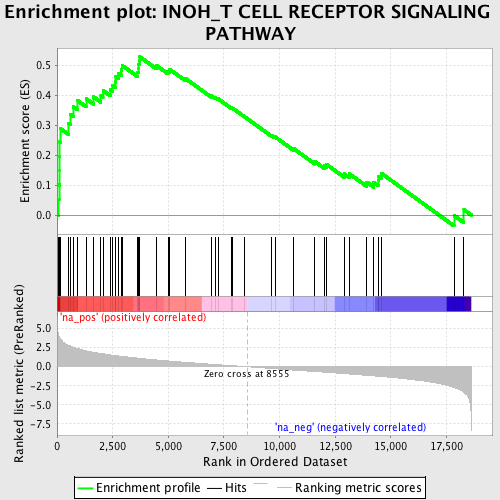
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | INOH_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.5288224 |
| Normalized Enrichment Score (NES) | 2.1321335 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.011856555 |
| FWER p-Value | 0.033 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TYK2 | 44 | 4.347 | 0.0537 | Yes | ||
| 2 | RAC2 | 88 | 3.901 | 0.1016 | Yes | ||
| 3 | SYK | 115 | 3.756 | 0.1486 | Yes | ||
| 4 | RASGRP1 | 118 | 3.738 | 0.1967 | Yes | ||
| 5 | ITPR3 | 123 | 3.720 | 0.2444 | Yes | ||
| 6 | BTK | 166 | 3.564 | 0.2881 | Yes | ||
| 7 | MAP2K1 | 502 | 2.825 | 0.3065 | Yes | ||
| 8 | JAK2 | 591 | 2.705 | 0.3366 | Yes | ||
| 9 | ITPR1 | 728 | 2.554 | 0.3622 | Yes | ||
| 10 | GRB2 | 924 | 2.371 | 0.3823 | Yes | ||
| 11 | MAPK3 | 1303 | 2.079 | 0.3887 | Yes | ||
| 12 | MAP2K2 | 1638 | 1.892 | 0.3951 | Yes | ||
| 13 | CSK | 1969 | 1.722 | 0.3995 | Yes | ||
| 14 | VAV1 | 2063 | 1.688 | 0.4163 | Yes | ||
| 15 | MAPK9 | 2391 | 1.543 | 0.4186 | Yes | ||
| 16 | PTK2 | 2477 | 1.514 | 0.4335 | Yes | ||
| 17 | SOS2 | 2609 | 1.463 | 0.4453 | Yes | ||
| 18 | FOSL1 | 2631 | 1.451 | 0.4629 | Yes | ||
| 19 | RAF1 | 2767 | 1.403 | 0.4737 | Yes | ||
| 20 | TEC | 2878 | 1.359 | 0.4853 | Yes | ||
| 21 | JAK3 | 2916 | 1.346 | 0.5006 | Yes | ||
| 22 | RELA | 3628 | 1.116 | 0.4767 | Yes | ||
| 23 | ARAF | 3649 | 1.111 | 0.4899 | Yes | ||
| 24 | JAK1 | 3653 | 1.108 | 0.5041 | Yes | ||
| 25 | NFKBIA | 3692 | 1.094 | 0.5161 | Yes | ||
| 26 | PLCG1 | 3717 | 1.086 | 0.5288 | Yes | ||
| 27 | PTK2B | 4470 | 0.877 | 0.4996 | No | ||
| 28 | RAC3 | 4987 | 0.751 | 0.4815 | No | ||
| 29 | NFKBIB | 5066 | 0.732 | 0.4868 | No | ||
| 30 | SOS1 | 5763 | 0.579 | 0.4567 | No | ||
| 31 | FOSB | 6919 | 0.321 | 0.3987 | No | ||
| 32 | FOS | 7117 | 0.279 | 0.3916 | No | ||
| 33 | NFKBIE | 7238 | 0.255 | 0.3885 | No | ||
| 34 | LAT | 7816 | 0.144 | 0.3593 | No | ||
| 35 | ITK | 7885 | 0.130 | 0.3573 | No | ||
| 36 | NFKB1 | 8403 | 0.033 | 0.3298 | No | ||
| 37 | NRAS | 9654 | -0.233 | 0.2655 | No | ||
| 38 | PRKCQ | 9793 | -0.264 | 0.2615 | No | ||
| 39 | KRAS | 10637 | -0.440 | 0.2218 | No | ||
| 40 | SRC | 11587 | -0.629 | 0.1787 | No | ||
| 41 | JUN | 12005 | -0.707 | 0.1654 | No | ||
| 42 | RAC1 | 12108 | -0.727 | 0.1693 | No | ||
| 43 | CD4 | 12899 | -0.899 | 0.1383 | No | ||
| 44 | HRAS | 13137 | -0.949 | 0.1378 | No | ||
| 45 | FYN | 13920 | -1.129 | 0.1102 | No | ||
| 46 | MAP2K4 | 14231 | -1.203 | 0.1090 | No | ||
| 47 | MAPK10 | 14428 | -1.253 | 0.1146 | No | ||
| 48 | ZAP70 | 14433 | -1.254 | 0.1306 | No | ||
| 49 | MAPK8 | 14563 | -1.284 | 0.1402 | No | ||
| 50 | LCK | 17843 | -2.721 | -0.0014 | No | ||
| 51 | FOSL2 | 18286 | -3.332 | 0.0178 | No |
